Nature: AI constructs gene interaction networks to accelerate discovery of disease treatment targets
The development of science and technology has made communication between people more and more convenient, with letters, emails, and social messaging software constructing the communication network of human society. And in our cells, there also exists a complex communication network connecting many genes - the gene network.
Mapping gene networks, that is, networks of interactions between different genes, is very important for understanding key biological and disease processes, as well as identifying potential drug targets. However, gene network mapping requires large amounts of transcriptomic data to infer connections between genes, which hinders discovery in data-limited environments, including rare diseases and clinically inaccessible tissues.
Recently, researchers from the Dana-Farber Cancer Institute in the US published a paper titled “Transfer learning enables predictions in network biology” in the journal Nature.
This study generated a gene expression dataset - Genecorpus-30M, which includes about 30 million single cell transcriptomic data from various human tissues. The research team pre-trained an AI model based on transfer learning - Geneformer, using this dataset to enable prediction of gene network dynamics, mapping of gene networks, and accelerated discovery of disease treatment candidate targets in limited data.
Fig.1
Today, artificial intelligence (AI) has shown great prowess in multiple fields and penetrated our daily lives. From AlphaGo in the game of Go to AlphaFold for protein structure prediction, from AI painting last year to ChatGPT today, as an emerging disruptive technology, AI is gradually unleashing the tremendous power accumulated from scientific revolutions and industrial changes, profoundly transforming human lives and mindsets.
In the field of biology, the application of AI is also expanding, such as AI therapeutics, AI drug discovery, and AI de novo protein design. Recently, a machine learning algorithm - transfer learning, has profoundly changed fields like natural language understanding and computer vision.
The working principle of transfer learning is to pre-train a deep learning model on large generic datasets, to perform a wide range of tasks with limited specific task data. In other words, transfer learning allows the fundamental knowledge obtained during pre-training to be transferred to tasks where the model has not yet been trained.
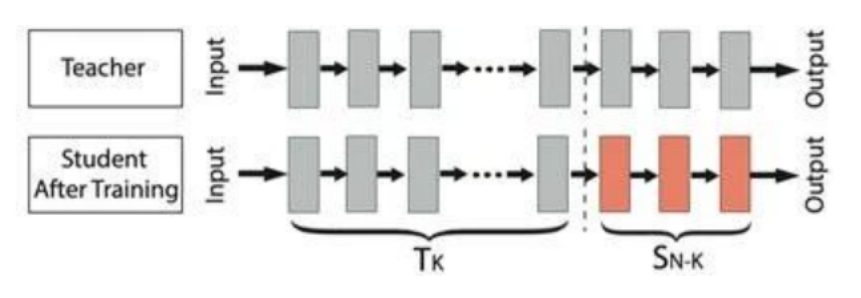
Fig.2 The core of transfer learning is "pre-training + fine-tuning"
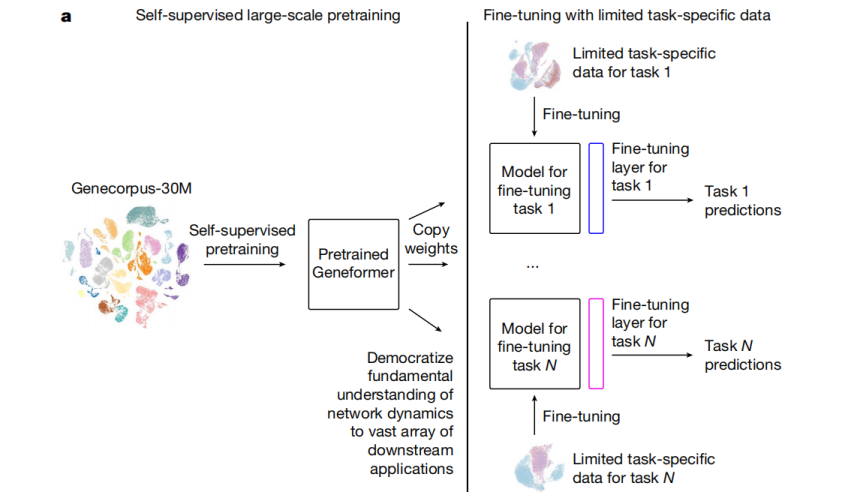
In this latest research, the research team set out to develop and pre-train a deep learning model with a large generic gene expression dataset, so that it could "understand" gene network dynamics, and provide predictions about gene interactions and cell states in a wide range of applications where data is scarce.
To achieve this, the research team first generated a gene expression dataset - Genecorpus-30M using public data, which included about 30 million single-cell transcriptomic data from a wide range of human body tissues. Then, the research team used this dataset to pre-train a deep learning model - Geneformer, developed based on transfer learning, to enable basic predictions about gene network dynamics.
Fig.3 The deep learning model called Geneformer, developed based on "transfer learning," that can predict gene network dynamics.
The pre-training of Geneformer is self-supervised, meaning it can learn from unlabeled data. As an attention-based model, Geneformer can learn which genes require more attention in the form of machine learning. Through this self-supervised pre-training, Geneformer learns to focus more on genes that play critical roles in the cell, such as genes encoding transcription factors and central regulatory nodes in gene networks.
Furthermore, Geneformer possesses environmental awareness, enabling it to make specific predictions based on the environment of each cell. This is crucial as gene functions differ across various cell types, developmental stages, and disease states. The research team states that Geneformer's environmental awareness is particularly useful for studying diseases and therapeutic targets affected by multiple cell types or progressive diseases that may vary across disease stages.
Fig.4 Geneformer improves the prediction of gene network dynamics with limited data.
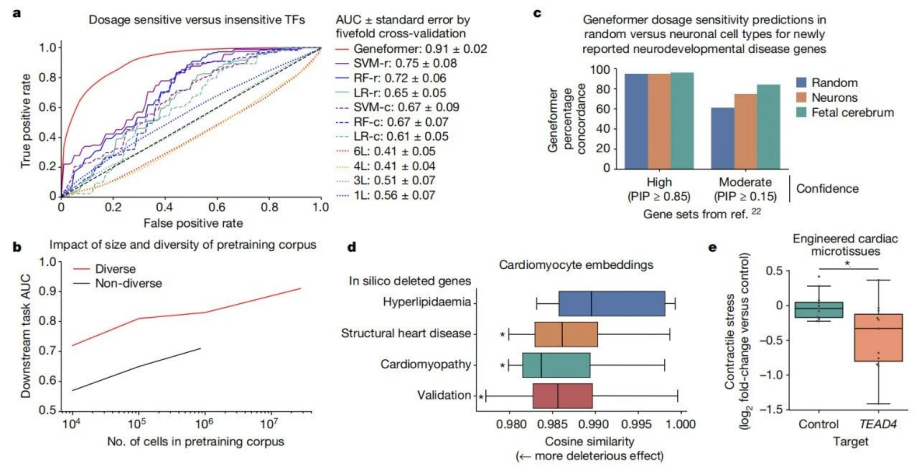
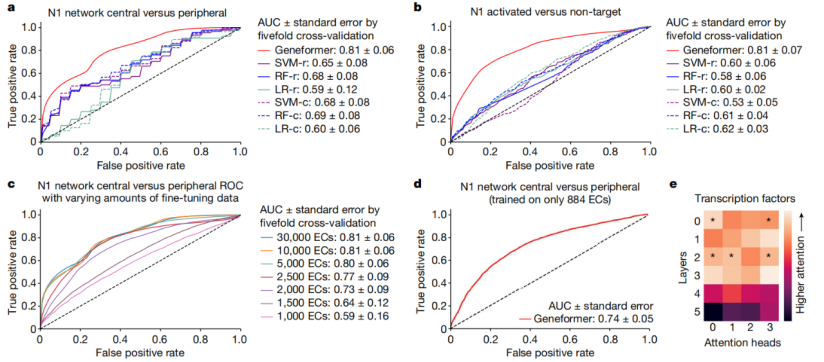
Pre-training allows Geneformer to encode the hierarchical structure of gene networks, including which genes influence the expression of other genes. The research team found that when Geneformer was fine-tuned for various tasks related to gene network dynamics or DNA-protein complex chromatin modifications, it consistently improved prediction accuracy compared to standard alternative methods.
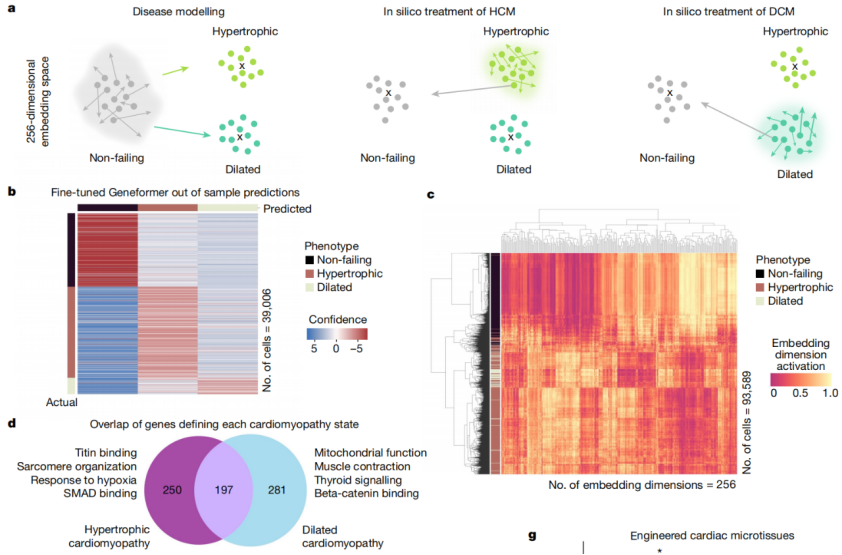
In this study, when fine-tuned using limited gene expression data specific to cardiac diseases, Geneformer identified candidate therapeutic targets. Targeting these candidates in induced pluripotent stem cell (iPSC)-based models of cardiac diseases resulted in improved contraction function of iPSC-derived cardiomyocytes.
Fig.5 Geneformer encodes the hierarchical structure of gene networks.
It is worth noting that pre-training Geneformer with larger and more diverse datasets can consistently improve its predictive ability in downstream tasks. This suggests that with the expansion of publicly available gene expression data, Geneformer can make accurate predictions even in more complex and ambiguous research contexts. For instance, research data on some rare diseases is often scarce, but Geneformer may be able to infer the pathological mechanisms of these rare diseases with just a small amount of data.
Fig.6 Geneformer reveals candidate therapeutic targets.
In summary, this study developed a deep learning model called Geneformer, based on transfer learning, which can depict gene networks. Through pre-training, fine-tuning, and transferring its understanding of gene network dynamics, Geneformer can be applied in various research fields to accelerate the discovery of key network regulators and candidate therapeutic targets using limited data.
With the increasing availability of multimodal data, future developments may involve creating deep learning models capable of jointly analyzing multiple types of data, such as mapping gene expression profiles and chromatin dynamics at the single-cell level. Additionally, future deep learning models can extract the network hierarchy encoded by Geneformer to infer specific cell types and disease network connections.
Paper link: