Favorite
RNA Splicer
AI-Powered RNA Splicing Prediction Tool
Please enter the mutation site in the following input box for prediction.
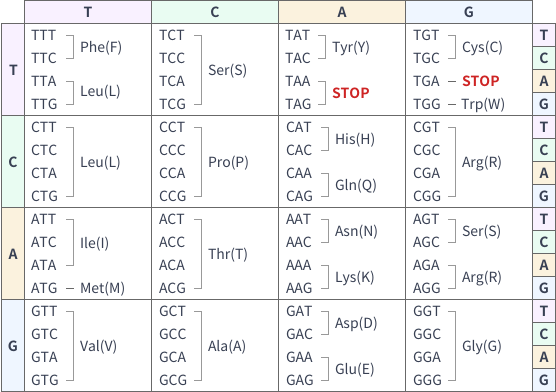
Codon Chart
Model Introduction
Tutorial (Video)
Tutorial (Text)
01
Why choose RNA Splicer?
Common challenges in RNA splicing analysis include:
- Determining whether a mutation affects RNA splicing
- Identifying exon skipping or retention
- Spending excessive time on literature review and manual analysis
RNA Splicer solves these issues with deep learning technology. By analyzing mutation data, it automatically predicts splicing pattern changes and generates clear visual results. Whether you’re conducting research, interpreting clinical findings, or preparing educational materials, RNA Splicer delivers accurate insights within minutes.
Key Highlights
Simple to Use
Enter a gene name and mutation information — the system automatically matches, analyzes, and predicts outcomes.
Visualized Results
Easily compare wild-type and mutant splicing patterns with clear, annotated graphics.
Multi-Species Support
In addition to human genes, RNA Splicer supports homologous mutation analysis in mice and rats.
Authoritative Standard
Complies with HGVS nomenclature for direct citation in both research and clinical settings.
AI Algorithm-Driven
Powered by deep learning for accurate and reproducible splicing predictions.
Step-by-Step Guide
Step 1: Open the Tool Interface
Step 2: Enter Mutation Details (Example: “USH2A gene c.8559-2A>G mutation”)
The tool supports four input modes: Paste, Mutation, Protein, and Transcript.
Step 3: Fill in Gene and Transcript Information (Example using Mutation)
- Enter the gene name (e.g., “USH2A”). The system will suggest available genes; select the target gene.
- Select the transcript: The system loads the most commonly used transcript by default (e.g., NM_206933.4 for USH2A). If there are special requirements, users can also select other transcripts from the dropdown menu.
Step 4: Input Mutation Site Information
- Fill in the mutation site:Enter "c.8559-2A>G" in the "Standard Mutation Nomenclature" box, and the system will automatically parse the pre-mutation base (A) and the post-mutation base (G).
- Visualize mutation site:Click the [Unfold Diagram] button below the input box to visually inspect the mutation's position within the gene structure (e.g., intron, exon boundary).
If you need to analyze multiple mutation sites, click the [Add Mutation] button (to the left of the input box) and repeat the above steps to add new mutations.
After confirming that the information is correct, click the [Submit] button at the bottom of the page to start the AI model prediction. Wait for the prediction results. A progress bar will be displayed on the page after submission; the prediction time depends on the number of mutations and server load.
Step 5: Interpret the Prediction Results
- Prediction Summary: Describes possible abnormal splicing outcomes (e.g., “123 bp deletion, exon skipping”).
- Splicing Diagram: Compares wild-type and mutant splicing sites, highlighting added/lost splice sites and exon changes.
- Numerical Score: Quantifies the impact on splicing (e.g., higher DanQ scores indicate greater likelihood of abnormal splicing).
- Cross-Species Analysis: Click [Mouse] or [Rat] to view predicted results for homologous mutations in other species.
Kind Reminder
Ensure gene names follow HGVS format and confirm transcript selection before submission. Up to 5 mutations can be submitted per analysis.
Try RNA Splicer now — make RNA splicing prediction effortless!
Please enter the mutation site in the following input box for prediction.
Codon Chart
Model Introduction
Tutorial (Video)
Tutorial (Text)
01
Interpretation of Results (Sample)
Prediction Output
Displays whether mutations may lead to abnormal splicing, such as “123 bp deletion, exon skipping.”
Visualization Diagram
Illustrates differences between wild-type and mutant splicing patterns, with clear annotations for altered splice sites or exon events.
Numerical Score
Provides a quantitative assessment of splice impact — higher scores indicate a higher probability of splicing alteration.
Cross-Species Comparison
View homologous mutation results for mice or rats with a single click on the result page.
Try RNA Splicer now — make RNA splicing prediction effortless!
Please enter the mutation site in the following input box for prediction.
Codon Chart
Model Introduction
Tutorial (Video)
Tutorial (Text)
01
FAQ / Q&A — RNA Splicing Prediction Model | AI-Powered Gene Mutation Analysis
Q1RNA 剪接预测模型是什么?
RNA Splicer 是一款基于深度学习的 AI 工具,能预测基因突变是否会导致 RNA 剪接异常。
Q2为什么选择 RNA Splicer?
操作简单、结果直观、支持多物种,科研与临床解读都可使用。
Q3是否需要安装?
不需要,打开网页即可直接使用。
Q4可以批量分析吗?
一次最多提交 5 个突变,批量研究时请分批输入。
Q5预测结果可靠吗?
工具基于深度学习模型(如 DanQ),结果附带量化分数,科研与临床研究均有参考价值。
Q6什么场景会用到RNA 剪接预测模型?
RNA Splicer serves research, clinical, and educational purposes. It rapidly verifies mutation effects on RNA splicing for mechanistic studies, aids in interpreting clinical genetic reports and assessing mutation pathogenicity, and visually demonstrates splicing mechanisms for teaching.
Other Useful Tools

Pathogenicity Predictor
AI tool for predicting the pathogenicity of gene mutations at any location
RNA Splicer
AI tool for predicting the impact of gene mutations at any location on RNA splicing sites
Sequence Alignment
Compare nucleotide or amino acid sequence similarities

Mutation Direct
Perform joint or cross searches for mutations, genes, and diseases