The user guide of ASO Designer
Introduction
ASO Designer is a software for predicting antisense nucleic acid (ASO) targets. It calculates the ASO targets in the query exon / input sequences plus its flanking 200bps. First, ASO Designer uses ViennaRNA and RNAstructure to calculate the binding energy of each possible ASO target, and then uses the poisson test to select the target with a statistically significant high binding energy.
Among the selected high binding energy targets, ASO Designer will calculate their other properties:
Melting point of ASO
Energy of ASO
Energy of ASO + ASO
GC percentage of ASO
The distance between the ASO target and the splicing acceptor
Whether the ASO target coincides with the enhancer and silencer of the query exon
Based on the above six characteristics, ASO Designer will rank the selected high binding energy targets, and the higher the ranking, the better the effect of ASO targets.
Instructions
Input information
The two prediction methods provided by ASO are genome prediction and gene prediction.
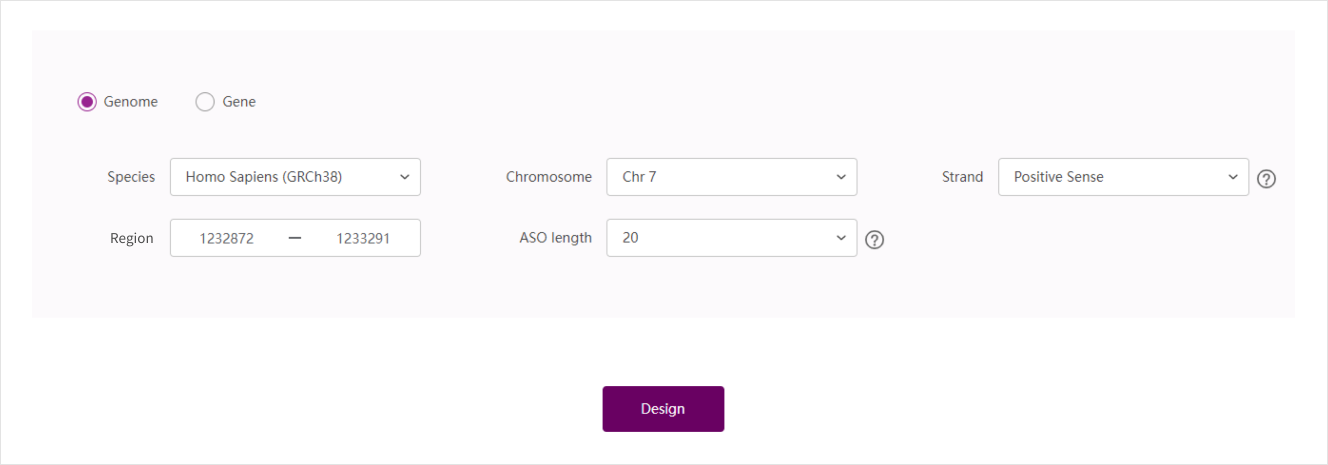
Genome prediction
Select species: ASO Designer currently only supports for two species, Human and Mouse. Among them, Homo Sapiens (GRCh37) and Homo Sapiens (GRCh38) represent the genome of the human GRCh37 version and the human GRCh38 version, respectively, and Mus Musculus (GRCm38) represents the genome of the mouse GRCm38 version.
Select chromosome: The chromosome number is used to accurately match the corresponding exon sequence. In biology, human chromosomes include 22 autosomes and two sex chromosomes X, Y, while mouse chromosomes only contain 19 autosomes and two sex chromosomes X, Y.
Select positive and negative strands: Due to the principle of complementary pairing between bases, a DNA strand is composed of two complementary DNA single strands. One of the single DNA strands is identical to its corresponding mRNA sequence, and this DNA strand is called the positive strand, and the other DNA strand is called the negative strand
Fill in the mutation position in the "Region" box
Choose ASO length: the length of ASO will affect the ASO effect. Empirically, ASO Designer provides ASO length between 20-25bps, which can perform a good effect.
Example: Homo Sapiens (GRCh38), chr7, positive chain, 1232872-1233291, 20
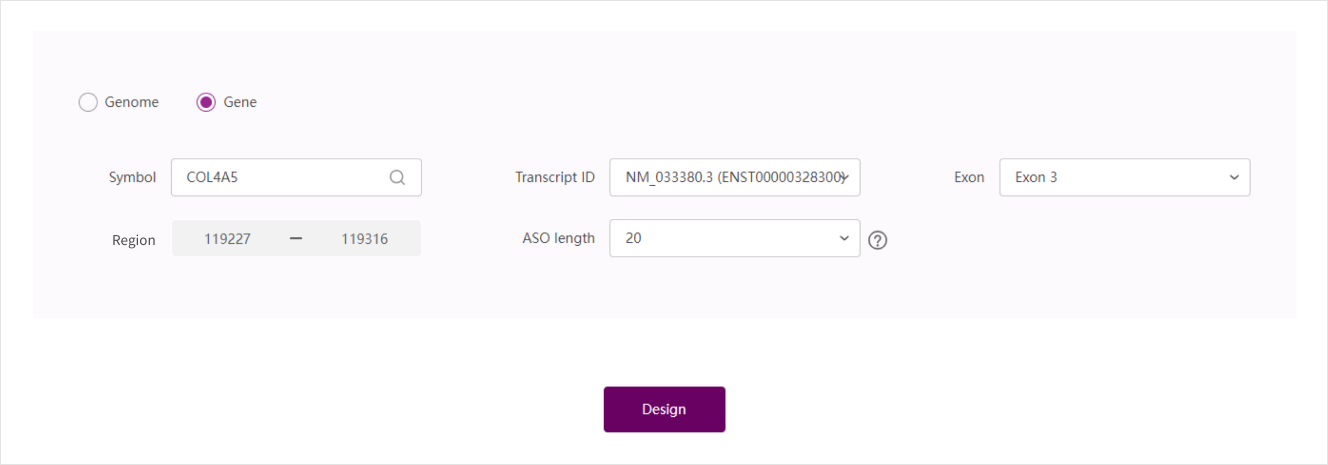
Gene prediction
Enter a gene for which you want to predict the ASO target
Select the corresponding transcript ID
Select the exon of ASO targets, and the system will automatically bring out the site of the exon in the "prediction region" (the site are the relative site of the gene)
Choose ASO length: the length of ASO will affect the ASO effect. Empirically, ASO Designer provides ASO length between 20-25bps, which can perform a good effect
Example: COL4A5, NM_033380.3(ENST00000328300), Exon 1, 1-369, 20
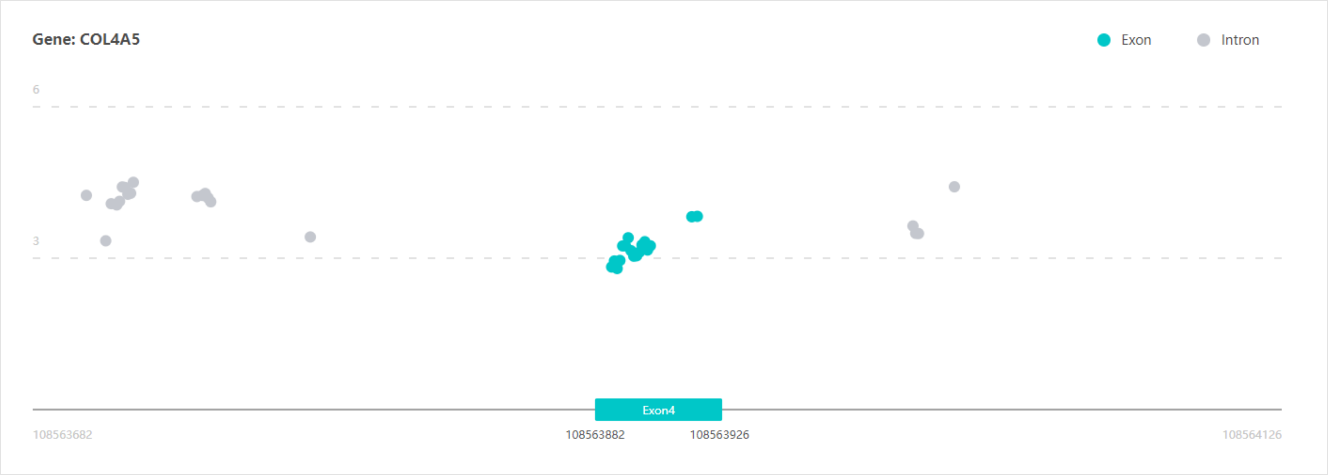
Prediction Results
Light blue represents the query exon/input sequence, and gray represents flanking 200 bp of the query exon / input sequence. Light blue dots represent possible ASO targets on query exon / input sequences, grey dots represent possible ASO targets on non-import sequences. The vertical axis represents the possible binding energy of ASO to the target.
X-axis: the site of query exon or input sequences
Y-axis: The binding energy of ASOs to the target
Intensity score: The sum of the normalized values of the melting point of ASO, the energy of ASO, the energy of ASO + ASO, the GC percentage of ASO and the distance of the ASO target from the splicing acceptor site
Start: The relative start site of the query exon/input sequence 200bps before and after
End: The relative end site of the query exon/input sequence 200bps before and after
OES: Does it coincide with the splice enhancer and silencer of the query exon







